The population genomics of herbicide resistance




The demographic and genetic origins of glyphosate resistance adaptation in Amaranthus tuberculatus
Harbouring cross-resistance to up to six classes of herbicides, the invasion of common waterhemp in agricultural environments is becoming increasingly problematic for crop productivity. With the weedy-agricultural waterhemp variety (Amaranthus tuberculatus var. rudis) having a range historically limited to the South-midwestern United States, first reports of waterhemp occurring in agricultural fields in Ontario, Canada have signalled the need to better understand the relative importance of the spread of resistant, weedy species, compared to the rapid adaptation of what were once benign populations (in this case, Amaranthus tuberculatus var. tuberculatus).
To this end, our work published in PNAS investigates how newly problematic agricultural populations arise, and how the spread of the species relates to the adaptive origins of glyphosate resistance. To dissect this question, we produced the first high quality reference genome for the species, characterized resistance and sequenced whole genomes from numerous populations spanning the epicentre of the agricultural invasion to much more recently established weed populations in Canada. We found that while for the most part, ancestry in these populations varied as expected with geography across the described range of the two subspecies, that populations in two key agricultural regions in Ontario, Walpole Island & Essex County, had drastically different evolutionary origins. Common Waterhemp individuals in Essex County appeared to be a collection of agricultural genotypes from the Midwestern United States—likely introduced through a seed dispersal event—while individuals in Walpole island much more closely resembled nearby natural populations.

These distinct demographic origins well predicted the genetic origins of glyphosate resistance. Genomes of individuals sampled from Walpole Island—in particular, patterns of similarity at a complex amplification of the gene targeted by round-up herbicides—pointed to glyphosate resistance spreading across this region from selection on a single de novo mutation. This fits with what we know about the recent and rapid conversion of this wetland habitat to agronomic practices, and naivety of this ancestry to such environments. In the midwest though, in populations that have persisted in agricultural environments on longer timescales and on the weedy var. rudis background which harbours higher genetic diversity, we find multiple independent origins of glyphosate resistance mutations. Several of these exact same mutational events also seem to have spread across the range up into Essex County, likely simultaneously with the original seed colonization event.
Kreiner JM, Giacomini D, Bemm F, Waithaka B, Regalado J, Lanz C, Hildebrandt J, Sikkema PH, Tranel PJ, Weigel D, Stinchcombe JR, & Wright SI. 2019. Multiple modes of convergent adaptation in the spread of glyphosate-resistant Amaranthus tuberculatus. PNAS. doi: 10.1073/pnas.1900870116
At the end of this project I still had a nagging question: How important is this gene amplification in conferring resistance to round-up herbicides across populations, relative to other (possibly unknown) genetic architectures?
The polygenic architecture of herbicide resistance
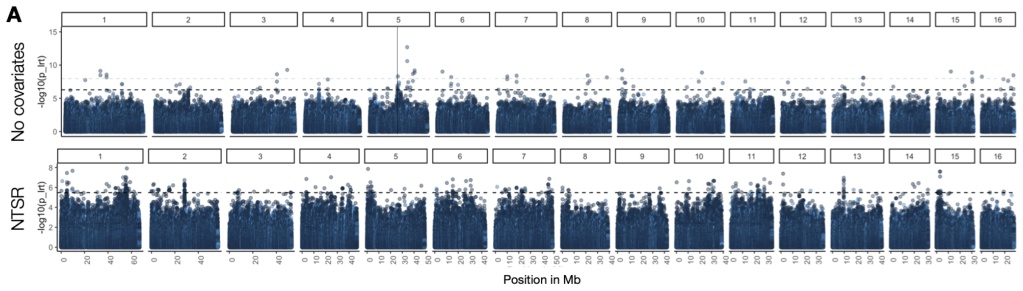
Mutations at genes that code for enzymes targeted by herbicides have by and large been the major focus of the study of herbicide resistance in weed populations. This tends to be because these causal mutations are easy to assay. For example, in our work on the adaptive origins of the amplification conferring glyphosate resistance (described above), I was able to focus my investigations to a very particular region of the genome. However, there are numerous other physiological mechanisms that confer resistance in natural populations—mechanisms for which we have no idea what loci and genes underlie this response. In our study populations of common waterhemp, a substantial portion of the variation in herbicide resistance is unexplained by known target-site mechanisms. In the map below, I’ve illustrated not only this point – that many individuals show resistance but lack known target-site mechanism – but the extent that the genetic architecture of resistance varies across populations.

I therefore set out to characterize the polygenic basis of resistance in common waterhemp, both with regard to genetic architecture of herbicide resistance (the number and distribution of genes involved, the frequency and effect size of resistance-loci) and how selection from herbicides has shaped genome-wide diversity. In particular, I was interested in how much variation in resistance across populations could be explained by genome-wide loci compared to monogenic mechanisms that are typically the focus of herbicide resistance research. In our paper at Molecular Ecology, published as a part of the special issue “Resistance evolution, from molecular mechanism to ecological context”, curated by Gina Baucom], we quantify the relative importance of polygenic vs monogenic herbicide resistance, show that strong selection shapes its genetic architecture with large effect & rare alleles, and find evidence of their maintenance by recent positive selection. Somewhat surprisingly, we show that polygenic resistance is of comparable importance to monogenic mechanisms in conferring round-up resistance in agricultural populations, even in individuals that already segregate for large-effect target-site mutations.
Kreiner JM, Tranel PJ, Weigel D, Stinchcombe JR, & Wright SI. 2021. The genetic architecture and population genomic signatures of glyphosate resistance. Molecular Ecology
The repeatability of simple target-site resistance evolution
Polygenic architectures and complex gene-amplifications are two very charismatic genetic underpinnings of herbicide resistance. Yet, even for the best studied genetic mechanism of herbicide resistance—simple single nucleotide polymorphisms—found to occur across hundreds of weed species, it remains unclear how often such mutations arise repeatedly within a single species; how long such mutations stick around in populations; the role of gene flow in their spread across the landscape; and how variable signatures of selection are across origins.
In a paper recently published in eLife, I characterized the repeatability of herbicide resistance evolution, using an increasingly tractable approach in population genomics, tree-sequence reconstruction, that takes into account not just the history of mutation but recombination. Not only did I find parallel evolution of herbicide resistance through changes at 11 distinct amino acids, but this tree reconstruction approach illuminates that 3 of these most common mutations themselves had arisen 7 times (A). By mapping these mutational origins back to the populations each lineage occurs in (B), I was able to decipher a key role of gene flow in the spread of resistance… nearly all mutational origins map to many populations, and most populations contain many mutational origins! Most of these mutational origins occurred after the onset of herbicide use, suggesting that variation for herbicide resistance is young and their persistence depends on herbicide use in these environments. Check out the article for more details on the strength of selection, recent population expansions, and the role of allelic interactions in shaping the distribution of resistance mutations across the range.
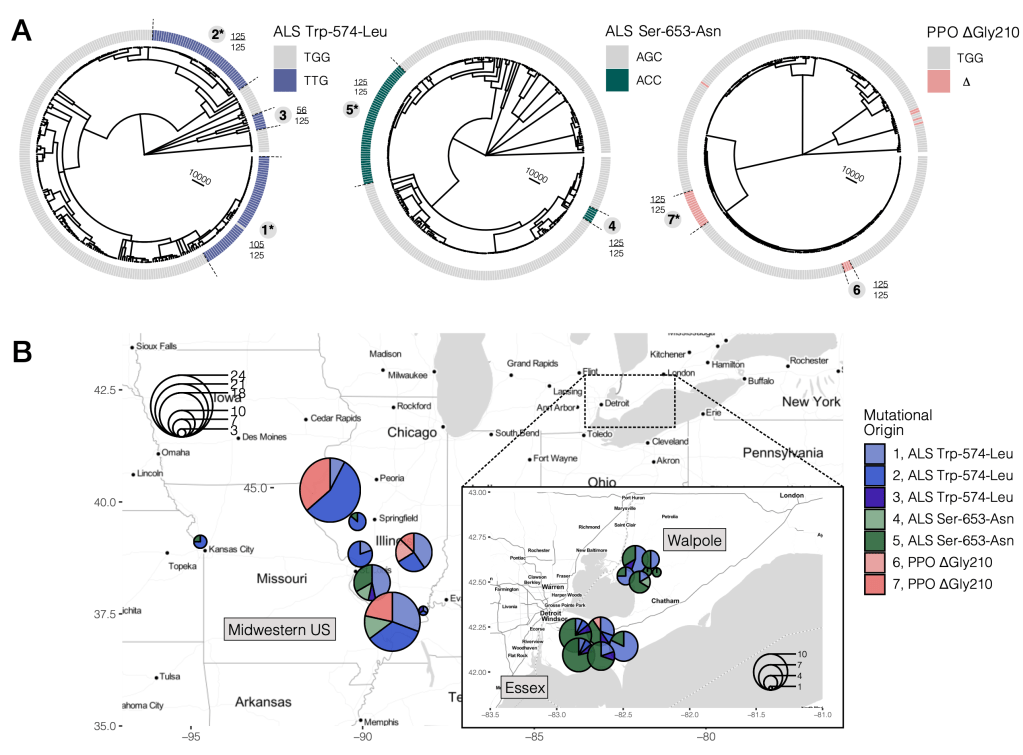
Kreiner JM, Sandler, G., Stern, A.J., Tranel PJ, Weigel D, Stinchcombe JR, & Wright SI. 2022. Repeated origins, gene flow, and allelic interactions of herbicide resistance mutations in a widespread agricultural weed. eLife. doi: https://elifesciences.org/articles/70242
